Enzymes

Roles of Enzymes
definition: highly specialized proteins that catalyze reactions in biological systems
-
decreases activation energy for a specific chemical reaction
-
are specific for one set of substrates
-
are not changed nor consumed in the reaction
nomenclature
-
usually ends with -ase
-
attached to substrate (urease) or description of action performed (lactate dehydrogenase)
-
sometimes have original names (trypsin)

6 Major classes of enzymes:
-
Oxidoreductases
-
catalyze REDOX reactions
-
ex. lactate dehydrogenase
-
-
Transferases
-
catalyze transfer of C,N or P containing groups
-
ex. serine hydroxylmethyl transferase
-
-
Hydrolases
-
catalyze cleavage of bonds by addition of water
-
ex. urease
-
-
Lyases
-
catalyze cleavage of C-C, C-S, and some C-N bonds
-
ex. pyruvate decarboxylase
-
-
Isomerases
-
catalyze racemization of isomers
-
ex. methylmalonyl CoA mutase
-
-
Ligases
-
catalyze formation of bonds between C, O, S and N coupled to hydrolysis of high energy phosphates
-
ex. pyruvate carboxylase
-
Molecular Structure
-
Enzyme molecular structure
-
Active site
-
Part of protein that where reactants come together
-
That way doesn’t rely on chance encounters
-
Arrangement of atoms into a 3-dimensional cleft or crevice
-
Substrate fits into the crevice—in most enzymes it’s an exact fit
-
“Lock and key”
-
-
Some enzymes: induced fit active site = active site is molded after the substrate engages
-
-
-
-
Enzymes are highly specific for their substrate
-
The reactant an enzyme acts on is the substrate
-
enzymes bind to substrates
-
enzyme-substrate complex
-
-
-
The reaction catalyzed by each enzyme is very specific.
-
results from its 3-dimensional shape, a consequence of its amino acid sequence.
-
-
The active site of an enzyme is typically a pocket or groove on the surface of the protein
-
As the substrate enters the active site, steric interactions between the chemical groups on the substrate and the R groups of amino acids of the protein cause the enzyme to change shape
-
-
This brings the chemical groups of the active site into position to catalyze the reaction.

k1
k2
k3
k4
Induced Fit Model

-
Active site approximately fits substrates (e.g., all proteins)
-
As substrate(s) begins to bind, conformation change in enzyme allows for better fit
-
Reaction takes place
study question:
-
how does the induced fit model explain enzyme-substrate specificity compared to the older lock and key model?

How do Enzymes Work?
-
Enzymes don’t work alone
-
Use coenzymes and cofactors
-
Cofactor (inorganic) e.g., Mg, Fe
-
not protein based
-
hold normal enzyme shape
-
allows substrate to bind to active site
-
-
Coenzyme (organic) e.g., vitamin based NAD, FAD
-
vitamin derived
-
does not directly interact with enzyme, is involved in reaction
-
used for multiple reactions
-
transfer small chemical groups from one reaction to another
-
-
-
-
Nomenclature
-
Holoenzyme = protein + cofactor (coenzyme)
-
Apoenzyme = protein alone (inactive)
-
-
This is the role of many vitamins in metabolism
study questions:
-
what 3 coenzymes are important in energy metabolism?
-
what is the main difference between cofactor and coenzyme?

Factors Affecting Enzymatic Rates
1. Catalytic Rate
-
Amount of product produced per unit time
-
Assumption: enzyme active site always occupied by substrate
2. Concentration
-
Greater enzyme concentration corresponds to greater reaction rate based on law of mass action
-
Greater substrate concentration – shows saturation (asymptotic response)
V

study questions:
-
how does substrate concentration affect enzymatic rate?
-
what occurs when enzymes are fully saturated?
-
number of collisions is dependent on the number of substrate molecules
-
reaction rate (V) is proportional to [S]
-
low [S] = first order reaction rate (linear)
-
-
when there is a large number of substrates, all enzymes cannot react at once
-
reaction rate increases and then plateaus at maximum rate/velocity (Vmax)
-
when product forms, more enzymes become available for substrate to react, creating equilibrium
-
high [S] = zero order reaction rate
-
3. Ligand-Protein Interactions (Affinity)
-
ligand - molecules that bind proteins by weak interactions
-
only hydrogen and ionic bonds, not covalent
-
affinity is the measure of strength of binding between substrate and enzyme
4. Acidity (pH)
5. Temperature

study question:
-
describe how affinity impacts reaction rate

Km, Vmax and Reaction Rates

What is Km?
-
Km is a substrate concentration
-
Units must be in concentration
-
A “constant” or defining feature of enzyme for a given set of conditions
-
-
Km is a property of every enzyme molecule…it does NOT depend upon the enzyme concentration. Thus it is an “intensive” constant, in contrast to Vmax.
-
Km is inversely related to the affinity of an enzyme for its substrate
-
the higher the affinity the lower the Km.
-

study question:
-
describe the relationship between Km and Vmax. how do they both impact enzyme rates?
-
lower Km means the enzyme has a higher affinity to the substrate
-
higher Km means the enzyme has a lower affinity to the substrate
-
Michaelis - Menton
-
Vi (initial velocity) = Vmax[S] / Km + [S]
-
Km = [S] at 1/2 Vmax
-
-
when [S] < Km
-
Lots of enzymes active sites are available
-
As [S], has strong effect on rate
-
First order kinetics--proportional to amount added
-
-
when [S] > Km
-
Because almost all active sites are occupied (saturated) there is limited amount of enzyme
-
More substrate does not affect rate more after reaction gets going
-
Zero order kinetics or independent of [S]
-


refer back to image below
In this scheme:
Km = (k2 + k3) / k1

k1
k2
k3
k4
-
Michaelis-Menten equation can be rearranged to linear form: Lineweaver-Burke
-
1/v = (Km/Vmax)(1/[S]) + 1/Vmax
-

Enzyme Inhibitors
-
Inhibitors are small molecules that bind to an enzyme and reduce its catalytic ability.
-
There are two major classes of inhibitors:
-
Reversible inhibitors can dissociate from the enzyme once they are bound
-
competitive, competitive, uncompetitive
-
-
Irreversible inhibitors can not dissociate from the enzyme.
-
Competitive
Competitive inhibitors react only with the free enzyme, often by binding to the active site…thus they “compete” with substrate for binding.


study question:
-
explain the differences between the 3 types of reversible inhibitors.




-
Inhibitor reversibly binds to the active site (where substrate would bind), therefore completing with substrate
-
reversed by increasing [S]
-
at sufficient [S], can still reach Vmax
-
increases Km for substrate
-
-
more substrate is needed to achieve 1/2 Vmax
-
the lineweaver-burke equation that describes the kinetics in the presence of a competitive inhibitor are altered by the term (1 + [I]/KI) as follows:
Non- Competitive
Non-competitive inhibitors react with the free enzyme and the enzyme substrate complex. Thus they usually bind to a site on the enzyme surface away from the active site.
-
decreased Vmax
-
occurs when inhibitor and substrate bind to different sites on the enzyme
-
can bind either the free enzyme of the ES complex and prevent reaction
-
cannot be overcome by increasing [S]
-
does not interfere with S binding to enzyme, no effect on Km
-
Vmax cannot be attained in the presence of a NC inhibitor
-
The lineweaver-burke equation that describes the kinetics in the presence of a non-competitive inhibitor are altered by the term (1 + [I]/KI) as follows:
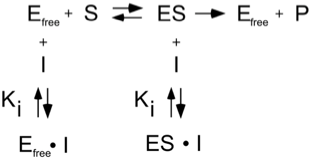


Uncompetitive
Uncompetitive inhibitors react only with the substrate bound form of the enzyme

The lineweaver-burke equation that describes the kinetics in the presence of an uncompetitive inhibitor are altered by the term (1 + [I]/KI) as follows:

Irreversible
-
Irreversible inhibitors inactivate enzymes by covalently binding them.
-
The kinetics of an irreversible inhibitor are quite easy to interpret: addition of inhibitor continually lowers Vmax until all enzyme molecules have reacted stoichiometrically with the inhibitor, at which point there will be no active enzyme molecules left.
-
Some irreversible inhibitors, called suicide substrates, look like the natural substrate but covalently attach to the enzyme at some point during binding and/or catalysis…high concentrations of substrate can temporarily protect the enzyme from such inhibitors.
Regulation of Enzyme Activity
1. Allosteric regulation
-
an enzyme has two binding sites: active and regulatory
-
modulator molecule binds to regulatory site which changes shape and activity of enzyme
-
can increase or decrease activity
-
generally alters affinity and catalytic rate
2. Covalent regulation
3. Feedback inhibition
4. Feedforward activation
